Simulating the spread of COVID-19 with BioDynaMo
Based on CERN’s experience in large-scale computing, BioDynaMo, an open-source computer simulation software, is now available in the European Open Science Cloud to support fighting the COVID-19 pandemic.

Animation of the spread of a virus
(in white: healthy; in red: infected; in blue: recovered)
In the wake of the COVID-19 pandemic, understanding how the virus spreads is crucial for governments when deciding on mobility restrictions, implementation of lockdown, and protective materials usage. Capable of simulating a large number of agents, platforms such as BioDynaMo could make a difference by sharpening our knowledge of crowd behaviour.
Originally designed to simulate the behaviour of billions of cells under specified environmental conditions, the BioDynaMo engine showed interesting potential for being adapted to run simulations of large human populations epidemiologists to model different epidemic scenarios. Within the scope of the CERN against COVID-19 task force, this open-source software could be modified to include elements of the dynamic spread of the SARS-CoV-2 virus spreads within a group of individuals.
First started as a CERN openlab project, BioDynaMo received seed funding from the CERN Medical Applications budget of 233 000 CHF over 5 years in order to improve its scalability and performances. At the peak of the first wave of the pandemic, the Medical Applications (MA) section in CERN’s Knowledge Transfer group further supported BioDynaMo. Its network enabled a collaboration with the University of Geneva’s Institute of Global Health for the implementation of a COVID-19 localised spreading model. Also, the MA section assisted the team in getting a grant from the European Open Science Cloud (EOSC) to develop the model yet more.
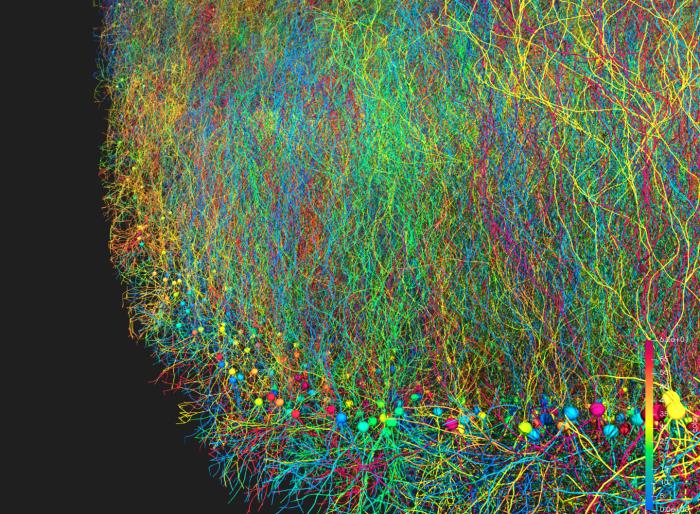
Large-scale simulation of neural development using BioDynaMo.
The project is expected to provide a valuable tool for helping the scientific community in their studies, with the goal of offering actionable insights to policymakers. Also, its clear visual representation can help underline the benefit of sensible social behaviour to prevent the spread of the disease.
Beyond the coronavirus pandemic, the know-how built during this project is useful for helping to design tools to contribute to preparedness for tackling similar viruses and future outbreaks.
Finally, the project will contribute to the EOSC Portal Catalogue and Marketplace by providing a service, openly accessible to the scientific community to predict the virus’ behaviour, and study how quarantines, confinements and social distancing influence the spreading panorama.
